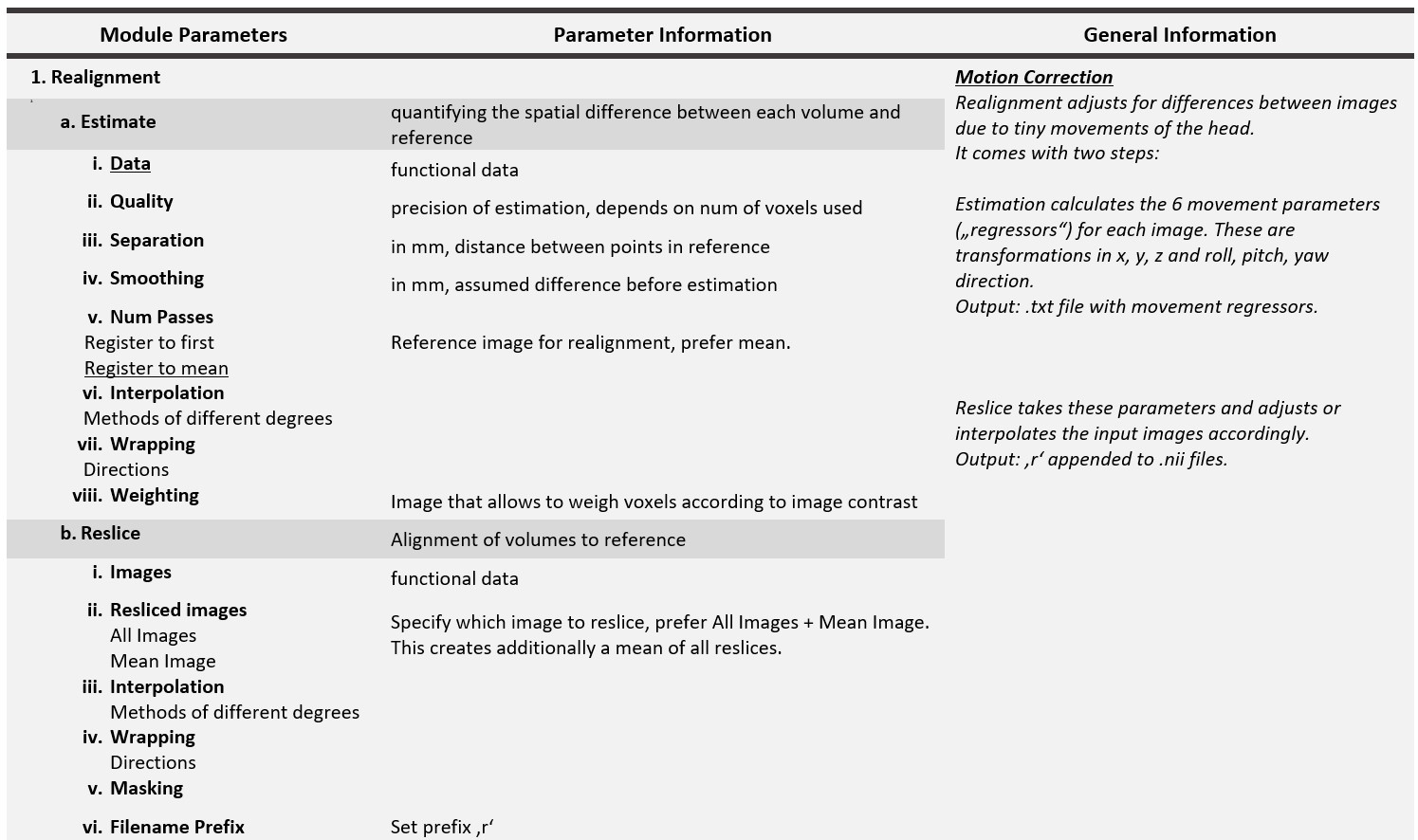
SPM Preprocessing
The SPM software allows you to customise preprocessing procedures by modulating the parameters and order of preprocessing steps. This has the advantage of allowing you to choose the most appropriate pipeline for your data, but also requires some initial understanding of the underlying analysis. Below is a list of all the main SPM preprocessing steps and their required parameters. Many of these parameters have a default value set by SPM. Do not change them if you do not understand their meaning. The underlined parameters have no default values and must be set in any case.
Motion Correction¶
Motion correction in SPM uses a 6-parameter based affine rigid body transformation in x, y, z, pitch, roll, yaw directions. The parameters are estimated by B-spline interpolation using a Taylor expansion (default: 4th degree) and minimising the sum of squared differences between a given functional image (first or mean) and all other images using Gauss-Newton optimisation. The B-spline interpolation approximates the sinc function, which is a good interpolation kernel except for sharp edges. The default estimation parameters (quality .75, fwhm 5, separation 4 and interpolation 4) are a good choice. They are even better than the more precise options (1.0 / 2 / 2 / 5)1.
Slice-Time Correction¶
 SPM applies the Fourier phase shift algorithm to the sinc-interpolated time series to correct for slice timing. To do this, it needs the time to repeat (TR) and the number of slices (N) to acquire a whole brain image. There are several ways in which the scanner can acquire the slices. Either the slice order is ascending
SPM applies the Fourier phase shift algorithm to the sinc-interpolated time series to correct for slice timing. To do this, it needs the time to repeat (TR) and the number of slices (N) to acquire a whole brain image. There are several ways in which the scanner can acquire the slices. Either the slice order is ascending [1:1:N], descending [N:-1:1] or interleaved.
If it is interleaved there are three more options:
&rarr middle slice to top
| Text Only | |
|---|---|
| Text Only | |
|---|---|
Coregistration¶
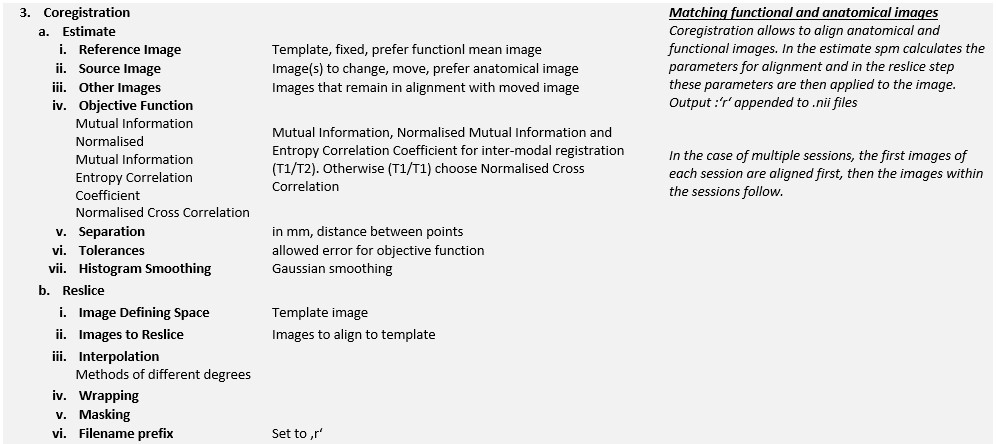 SPM cannot use minimum squared differences to align the anatomical and functional images. This is because the T1 and T2 images have different image properties, where the grey scales are opposite to each other. Therefore, the mutual information criterion is commonly used to measure the differences between the two image types. Again, the standard interpolation method is used with optimal parameters for estimation and processing time.
SPM cannot use minimum squared differences to align the anatomical and functional images. This is because the T1 and T2 images have different image properties, where the grey scales are opposite to each other. Therefore, the mutual information criterion is commonly used to measure the differences between the two image types. Again, the standard interpolation method is used with optimal parameters for estimation and processing time.
When coregistering the anatomical and functional images, the structural image is often normalised because it has a higher spatial resolution and fewer artefacts and distortions. Furthermore, co-registration is often only estimated, not applied, as this would reduce image quality. The estimated parameters are then used to transform the functional images for presentation purposes.
Segmentation¶
 SPM segmentation uses the Unified Segmentation approach 2 to combine segmentation, bias correction and spatial normalisation into one model.
SPM segmentation uses the Unified Segmentation approach 2 to combine segmentation, bias correction and spatial normalisation into one model.
It classifies tissue into white matter, grey matter, cerebrospinal fluid (CSF), bone, soft tissue and air/background. This helps to normalise the images later. At the same time, it removes uneven distortions in signal intensity. To do this, SPM uses tissue probability maps, templates that are aligned with the data to assign specific tissue types to voxels.
The tissue maps can be stored as warped (normalised) tissue, either unmodulated or modulated, which removes changes in image intensities over time. This is particularly important for MRI data as image intensities change.
Normalize¶
 SPMs normalization algorithm assumes that the images are aligned along the anterior commisure posterior commissure line (AC-PC). It might be useful to realign the images using a simple script or check them visually beforehand.
SPMs normalization algorithm assumes that the images are aligned along the anterior commisure posterior commissure line (AC-PC). It might be useful to realign the images using a simple script or check them visually beforehand.
Check normalization (Check reg button) afterwards. If normalization failed (more than 3cm or 15 degrees misalignment) then reposition the images via Display button.
Smoothing¶
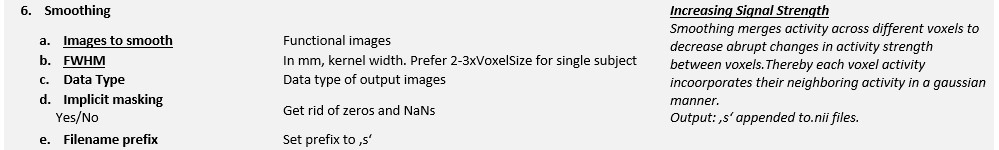 SPM applies a 3D Gaussian Filter across each voxel to smooth the images. The width of the kernel is defined by the FWHM and is commonly set between 8-10 mm.
SPM applies a 3D Gaussian Filter across each voxel to smooth the images. The width of the kernel is defined by the FWHM and is commonly set between 8-10 mm.
-
Oakes, T. R., Johnstone, T., Walsh, K. O., Greischar, L. L., Alexander, A. L., Fox, A. S., & Davidson, R. J. (2005). Comparison of fMRI motion correction software tools. Neuroimage, 28(3), 529-543. ↩
-
Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005 Jul 1;26(3):839-51. doi: 10.1016/j.neuroimage.2005.02.018. Epub 2005 Apr 1. PMID: 15955494. ↩